可视化案例
在生物信息学研究中,数据可视化是理解和解释数据的关键步骤。通过有效的可视化,研究人员可以更直观地展示数据的趋势、模式和关系,从而获得更深入的生物学见解。在这一部分,记录一些学习工作中R语言可视化案例。
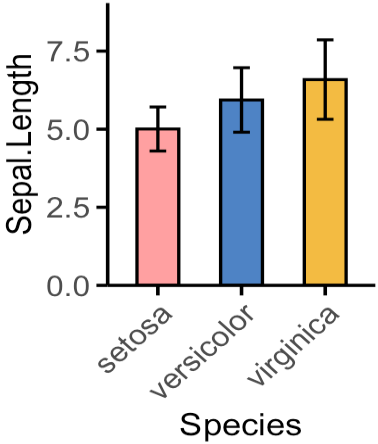
柱状图(Bar Plot)
df <- as_tibble(iris)
mycols <- c("#ffa0a1", "#4d83c5", "#f3bb42")
df %>%
ggplot(aes(x = Species, y = Sepal.Length, fill = Species)) +
geom_bar(stat = "summary",
fun = mean, # 计算均值
size = 0.6,
width = 0.5, # 设置柱状图宽度
color = "black") + # 设置柱状图边框颜色
stat_summary(fun.data = mean_sdl, # 添加均值标准差误差线
geom = "errorbar",
width = 0.2,
size = 0.6) +
scale_fill_manual(values = mycols) + # 应用自定义颜色
scale_y_continuous(expand = expansion(mult = c(0, 0.15))) + # 控制 Y 轴扩展,使Y 轴底部没有扩展空间,顶部有 15% 的数据范围作为扩展。
theme_classic() + # 使用经典主题
theme(legend.position = "none", # 隐藏图例
axis.title = element_text(size = 13), # 设置坐标轴标题大小
axis.text = element_text(size = 12), # 设置坐标轴文本大小
axis.ticks.length = unit(0.15, "cm"), # 设置刻度长度
axis.line = element_line(size = 0.65, color = "black"), # 设置轴线样式
axis.ticks = element_line(size = 0.65, color = "black"), # 设置刻度线样式
axis.text.x = element_text(angle = 45, hjust = 1))
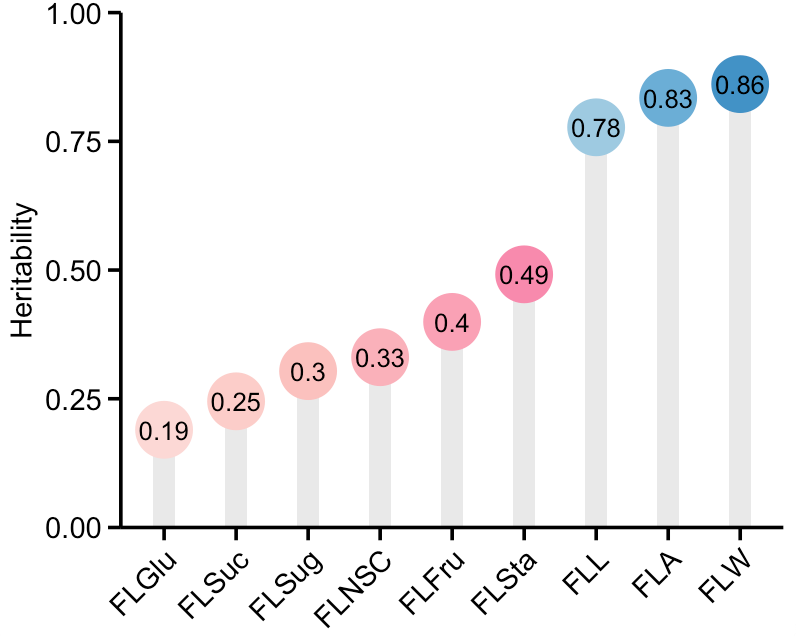
df <- read_csv("~/Desktop/Supplemental_Table_1.csv")
# 根据Heritability的大小对Traits进行排序
df <- df %>%
arrange(Heritability) %>%
mutate(Traits = factor(Traits, levels = unique(Traits)))
df %>%
ggplot(aes(x = Traits, y = Heritability)) +
geom_bar(stat = "identity", fill = "#E9E9E9",width = 0.3) +
geom_count(aes(color = Traits),size = 9.5) +
geom_text(aes(label = round(Heritability, 2)),size = 3.4, hjust = 0.5) +
labs(x = NULL) +
scale_y_continuous(expand = c(0,0),limits = c(0,1)) +
scale_x_discrete(labels = c("FLGlu", "FLSuc", "FLSug", "FLNSC", "FLFru", "FLSta","FLL","FLA","FLW")) +
scale_color_manual(values = c("#FCD8D5", "#FCCDC9", "#FBC1BE", "#FAB1BA",
"#FAA1B5", "#F88AAD", "#9ecae1","#6baed6", "#4292c6")) +
theme_pubr(base_size = 11) +
theme(legend.position = "none",
axis.text.x = element_text(angle = 45, hjust = 1),
axis.ticks.length = unit(0.17, "cm"),
axis.line = element_line(size = 0.65, color = "black"),
axis.ticks = element_line(size = 0.65, color = "black"),)
ggsave("~/Desktop/Heritability.pdf",width = 5, height = 3.8)
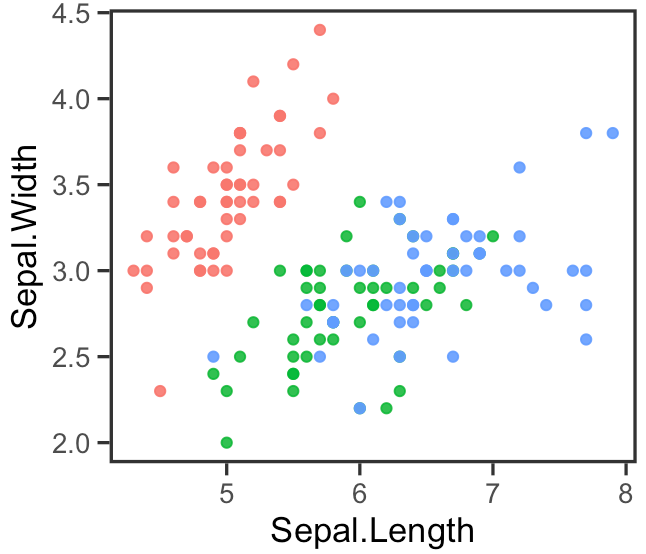
散点图(Scatter Plot)
# 加载必需的库
library(ggplot2)
library(dplyr)
# 使用管道操作符 %>% 来处理数据和创建图形
df %>%
ggplot(aes(x = Sepal.Length, y = Sepal.Width, color = Species)) + # 设置aes映射,定义x、y轴数据和颜色分组
geom_point(size = 1.5, alpha = 0.9) + # 添加点图层,设置点的大小和透明度
theme_test(base_size = 13, base_rect_size = 1.2) + # 使用theme_test主题,调整基本字体大小和矩形大小
theme(legend.position = "none", # 隐藏图例
axis.ticks.length = unit(0.16, "cm")) # 设置坐标轴刻度的长度
火山图(Volcano Plot)
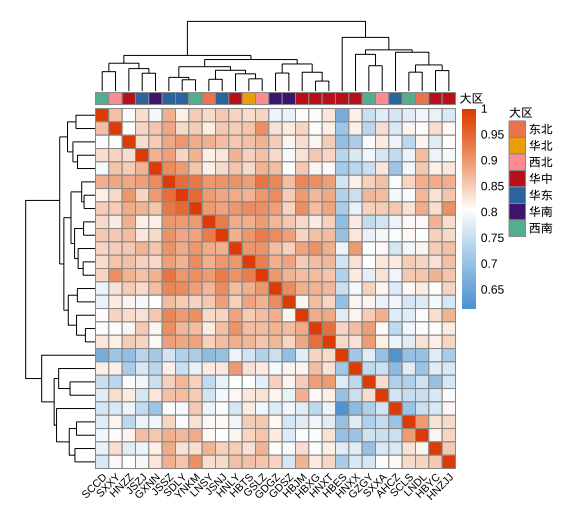
热图(Heatmap)
样品相关性矩阵热图示例数据
library(tidyverse)
library(pheatmap)
library(RColorBrewer)
library(showtext)
showtext_auto() # for Chinese characters
加载数据
data <- read.csv(url.1, header = T,row.names = 1)
# transform the data to a matrix
data_mt <- as.matrix(data)
# columns ann file
ann_col <- read.csv(url.2, row.names = 1)
ann_colors <- list(大区 = c(东北="#f08961", 华北="#f0ad00", 西北="#ffa0a1",
华中="#c7231f", 华东="#3578ad",
华南 = "#4f2580", 西南="#63b99e"))
# Set the color for the heatmap
mycols <- colorRampPalette(c("#5fa5d9", "white", "#e65100"))(100)
Draw heatmap
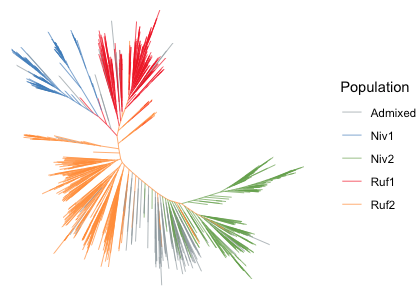
进化树
library(ggtree)
library(ggplot2)
library(RColorBrewer)
clist <- list(
"mycolor"=c("#97A1A7FF","#4d83c5","#74ab4d","#e61f19","#fe9929")
"shiny"=c("#1D72F5","#DF0101","#77CE61", "#FF9326","#A945FF","#0089B2","#FDF060",
"#FFA6B2","#BFF217","#60D5FD","#CC1577","#F2B950","#7FB21D","#EC496F",
"#326397","#B26314","#027368","#A4A4A4","#610B5E"),
"strong"=c("#11A4C8","#63C2C5","#1D4F9F","#0C516D","#2A2771","#396D35","#80C342",
"#725DA8","#B62025","#ED2224","#ED1943","#ED3995","#7E277C",
"#F7EC16","#F8941E","#8C2A1C","#808080"))
# 读取文件
groupfile <- read.table("group.tsv",sep = '\t',header = T)
tree <- read.tree("iqtree.tree")
# 准备颜色分组
grouplist <- split(groupfile$Sample, groupfile[,"Group"])
tree <- groupOTU(tree, grouplist, "Population")
ggtree(tree, aes(color=Population), alpha=0.8,layout='equal_angle',size=0.3) +
coord_flip() +
scale_color_manual(values = c("#97A1A7FF","#4d83c5","#74ab4d","#e61f19","#fe9929")) +
theme(legend.position='right')